Fat and the Hippo Pathway
Fat and Dachsous (Ds) are extremely large cell-cell adhesion molecules, first identified in Drosophila, that function in growth control and a form of tissue organization known as Planar Cell Polarity (PCP). Drosophila Fat possesses 34 cadherin repeats and has a molecular weight of 560kDa, while Ds has 27 cadherin repeats and a molecular weight of 350kDa. The extracellular domain of Ds binds to the extracellular domain of Fat. Together, Fat and Ds regulate cell proliferation through the Hippo kinase pathway. Ds binding to Fat regulates Fat’s activity in PCP and in the Hippo pathway. Ds is expressed in a gradient in tissues that display PCP, directing PCP and possibly growth control. Fat and Ds regulate expression of Four-jointed (Fj) (a Golgi-associated kinase) and other, as yet unidentified PCP target genes via the transcriptional co-repressor Atrophin. Planar polarity is also regulated by the core PCP pathway.. There are four Fat genes in mammals (Fat1-4); all have 34 cadherin repeats, EGF domains and Laminin G motifs in their extracellular domain, with divergent intracellular domains. Fat4 is the closest ortholog of Drosophila Fat, while Fats1-3 are more similar to another Drosophila cadherin called Fat-like. We showed that the Fat4 regulates PCP in mouse models, and that loss of Fat4 leads to cystic kidney disease. However, the biochemical mechanisms by which Fat regulates PCP in mouse or flies is still unclear.
Hippo Pathway
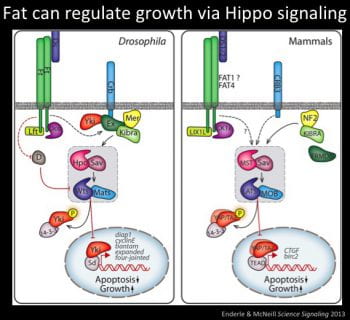
The Hippo Pathway is a highly conserved growth pathway, consisting of a kinase cascade that regulates growth via phosphorylation of a transcriptional co-activator called Yorkie (YAP/TAZ in mammals). We showed that Drosophila fat functions upstream of the Hippo pathway to regulate proliferation and apoptosis. The mammalian Hippo homolog, MST, phosphorylates and activates the Warts orthologs Lats1 and Lats2, which in turn phosphorylates and suppresses YAP/TAZ. Analysis of public database supports the proposal that Fat cadherins are involved in cancer; 16% of all tumors reported in the cBioportal have mutations in Fat cadherins. However the causal link between Fat cadherins and cancer is still unclear, and how it could regulate Hippo pathway activity is still unknown.
Research Approach
Our research approach combines biochemistry and molecular biology with genetics and high-resolution imaging to determine how tissue organization and proliferation are coordinately controlled in vivo during development.
Use biochemical approaches to further dissect Fat cadherin function.
We are using proteomic screening to identify Fat cadherin pathway effectors with mass spectrometry (MS). Using MS we have identified proteins that bind the cytoplasmic domain of Fat4, and found binding partners that have the potential to explain some of the effects of loss of Fat4 in PCP and Hippo activity. We will extend this approach to Drosophila Fat and Ds, as well as all mammalian Fat and Dachsous cadherins. Co-culture assays will determine how Fat-Dachsous binding affects the profile of proteins that bind the cytoplasmic domain of each protein. MS analysis of components of the Fat/Ds pathway will identify changes in post-translational modifications upon Fat-Ds binding. Interacting domains will be defined via co-immunoprecipitation experiments and direct interactions assayed with purified proteins.
Use Drosophila genetics to clarify the Fat signaling pathway, test the relevance of interactors found in biochemical screens, and identify new pathway components.
We will use Drosophila to explore the role of conserved Fat/Fat4 pathway protein interactors identified in our AP-MS experiments, as Fat pathway phenotypes are rapidly and easily analyzed in flies and PCP cannot be effectively assayed in tissue culture. We will use mutant alleles or transgenic RNAi to explore the function of Fat4 binding proteins. Proteins that show altered interactions dependent on Fat-Ds binding, or binding regulated posttranslational modifications will also be prioritized for in vivo analysis. Protein interactors that our in vivo analysis in Drosophila indicates are involved in Hippo or PCP will be explored in further biochemical studies. We will conduct genetic screens in Drosophila in a sensitized background (fat hypomorphic alleles) to identify new Fat pathway components. CRISPR will be used to mutate binding residues in Fat to dissect their contribution in vivo.